Date and time manipulations with CDO
Several tasks relating to manipulating netCDF files and dates/times within them are noted here.
- Converting the units and values of the
timevariable.
Setting the time ‘axis’
I have a netcdf file that contains a time variable. The values in the variable’s data array represent time units since date time string. Some model output and data formats use months since as the unit, which strictly speaking is non-standard and not CF compliant. (The integer values in the time variable array simply count increasing months, for example.)
Instead, I would like these to be represented as days since. So the integer values for time should now represent months.
I also want the month value to be reperesented at the mid-point of the month (The 15th), so I have chained in the -settaxis command as well.
cdo settunits,days -settaxis,2001-01-15,00:00,1month INFILE.nc NEW_OUTFILE.nc
The command settunits will change the time:units value as well, if it is in standard CF format.
It is useful to check the output is correct using cdo showdate or cdo showdatetime
2001-01-15 2001-02-15 2001-03-15 2001-04-15 2001-05-15....
Why do Python functions return None?
I was talking to a colleague recently about what would be covered in a basic ‘introduction to python’ tutorial. We got to talking about lists and how you would explain how to add things to lists with the append() method.
We had this code that turned out to be wrong because we’d forgotten that list.append() does not actually return a list, it merely updates the list object in place. (I.e. there is no return value from the function. Other programming languages might call this a void function or a procedure.
What we tried to do was:
l = ['one', 'two', 'three']
print(l.append('four'))
And to our (initial) surprise, got:
None
Instead we should have called the method first (updating the list, l) and then called print(l) to see the updated list.
Inspecting our mistake, we investigated what the type of l.append(item) was:
type(l.append("four")
# NoneType
Yep, definitely None it seems! But why is it None? If it has no return value, why does it have any type at all, why not raise an error?
What about other functions with no return statement, we thought…
def voidfunc(a, b):
x = a + b
type(voidfunc(1, 2)
# NoneType (?!)
z = voidfunc(1, 2)
print(z)
# None
This last one is slightly confusing, because you might intuitively expect an error perhaps (how could you assign a value to a variable from a function with no return statement?)
In fact, it turned out from some googling that Python functions have a default return value, which is None if no return expression is given, or return is given on its own.
For example, the source code for the append() method of list is this:
In the CPython source, this is in
Objects/listobject.c
static PyObject *
list_append(PyListObject *self, PyObject *object)
{
if (app1(self, object) == 0)
Py_RETURN_NONE;
return NULL;
}
It actually calls another function app1() which does the actual appending, which returns 0 on success. The next line Py_RETURN_NONE gives us a hint to what happens in the append method.
The builtins are a little confusing, so I’ll come back to them another time, but here is the code which determines what happens in functions that we write ourselves:
Python/compile.c
/* Make sure every block that falls off the end returns None.
XXX NEXT_BLOCK() isn't quite right, because if the last
block ends with a jump or return b_next shouldn't set.
*/
if (!c->u->u_curblock->b_return) {
NEXT_BLOCK(c);
if (addNone)
ADDOP_LOAD_CONST(c, Py_None);
ADDOP(c, RETURN_VALUE);
}
Without going in to the details of this code, there is a conditional statement that makes sure to “add None” if there is no return value given. (Py_None is the Python None object, see https://docs.python.org/2/c-api/none.html)
Masking variables in netCDF files with a landsea mask
These notes record how to mask (set to a no-data or missing-data value) a single netCDF variable, using a landsea mask.
Problem: We have a netCDF file containing a variable mapped over the globe. The variable should not have any values over the sea areas, because that doesn’t make sense for this particular data. (Let’s say it’s output from some post processed satellite data for argument). All the data points over seas and oceans are zero, but we can’t use zero as ‘NoData’ becuase zero is a valid value over land as well.
We want to use the _FillValue attribute of netCDF variables as a mask, so that when plotting or calculations are done using the variable, the sea values are ignored.
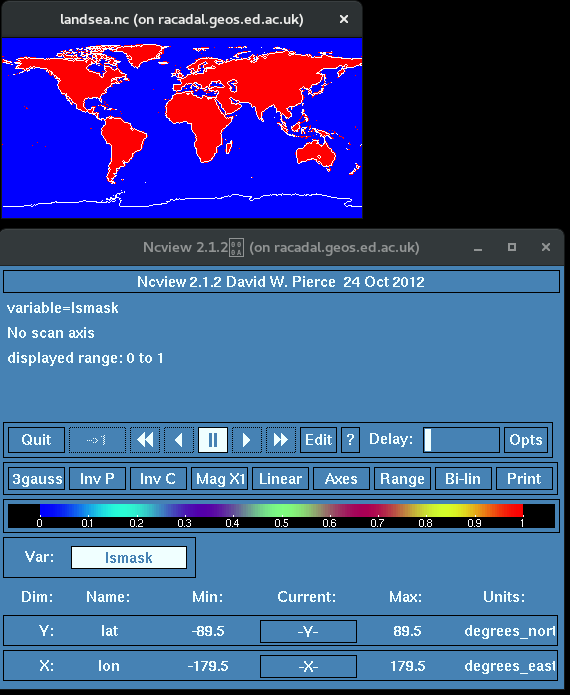
We also have a binary 1/0 land-sea mask in a separate file, at the same resolution as out JULES out put data. The land values have 1, and the seas/oceans have 0.
Input files
landsea.nc
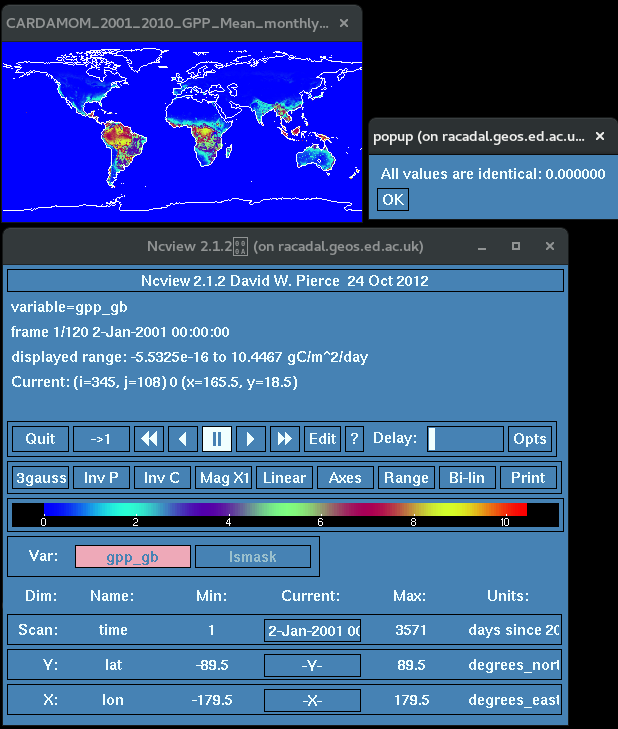
data_to_be_masked.nc
Steps
-
First copy over the landsea mask variable from landsea.nc to the file to be masked
ncks -A -v lsmask landsea.nc data_to_be_masked.nc -
Then make sure that the variable in the other file (gpp_gb) has a _FillValue set correctly (using -9999 here, but any convention will do (not zero though…)
ncatted -a _FillValue,gpp_gb,o,f,-9999 data_to_be_masked.nc -
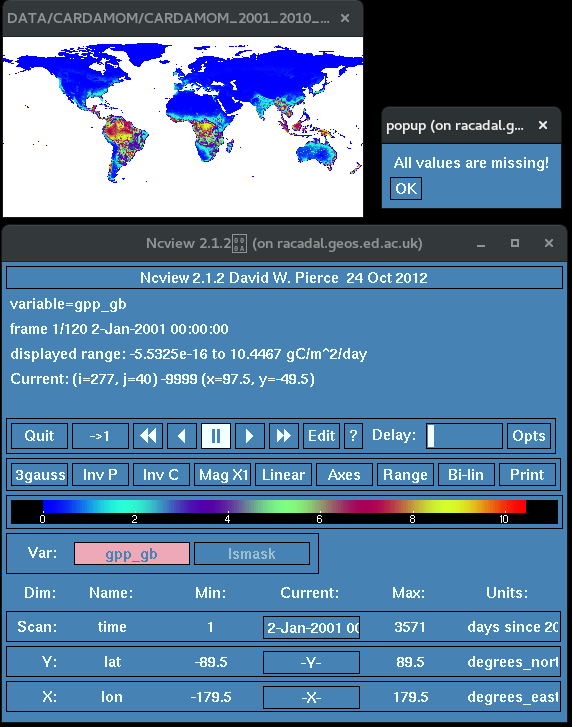
Then use ncap2 to set all the gpp values to _FillValue (the mask) whereever the landsea mask is not land (i.e. not == 1)
This is written to a new file.
```
ncap2 -s 'where(lsmask != 1) gpp_gb=gpp_gb@_FillValue' data_to_be_masked.nc data_after_masking_done.nc
```
The output variable gpp_gb now has the sea values set to the Nodata FillValue.
Deleting and adding attributes and variables with NCO
Here are some more common tasks I’ve come across when needing to edit netCDF files. This is usually when they need to be ingested into different models or post-processing scripts that require the netCDF files to be in a certain format.
Deleting a global attribute
You want to delete a single global attribute from a netCDF file.
This can be done using ncatted, e.g.:
ncatted -a global_attr_name,d,, infile.nc outfile.nc
This command takes for arguments separated by commas. Since we are specifying deletion, (d), only the first two arguments are needed, but the remaining commas bust be typed in.
Convert a variable type
A variable is of incorrect type and you need to change it. You can use ncap2 (nc arithmetic processing).
ncap2 -s 'time=float(time)'
Assumes you already have the variable defined. The -s option specifies that we are providing an inline script, within the quote marks.
Add a variable mapped over a certain dimension
You want to a variable that iterates over a given dimension, such as time. The variable should increase montonically (i.e. increase by n each time until the end of the dimension length is reached. I often find I need to do this after having merged netCDF files that were single time slices from a model output or satellite data or otherwise. ncap2 is used.
ncap2 -s 'time[$time]=array(54760,30,$time)' infile.nc outfile.nc
We are assigning the current time variable (assuming we have already added this) an array of values, specified by the (start_point, step, dimension). In this case, we get an array of values starting at 54760, increasing by 30 each point, as long as the time dimension. The -s option simply means we are giving an inline script as the input to the ncap2 program.
Add an attribute one at a time
You want to an attribute to a variable. (I.e. metadata attributes for variables, such as units, etc.). We can use ncatted for this. (netCDF attribute editor).
ncatted -a attribute,variable,a,c,"Atrribute Value" infile.nc
The -a option specifies append mode, and so we only need to supply the input file infile.nc. The value of the attribute is given in the quotation marks. The nco documentation suggested also putting single quotation marks around the comma-separated arguments as well, but I found this produced unexpected results where the double quotes were escaped and inserted into the actual attribute value as well. Could possibly be a unix thing though…
A netcdf bug to watch out for…
Merging netCDF files with NCO/CDO
NCO (NetCDF Operators)
You can merge netcdf files with the nco package. NCO is a set of linux command line utilities for performing common operations on netcdf files. This is useful for mergeing a set of files such as:
ModelRun_Jan.nc
ModelRun_Feb.nc
ModelRun_Mar.nc
...
Concatenating files, creating a new dimension in the process
To concatenate files, use ncecat:
ncecat *nc -O merged.nc
This will merge all the netcdf files in a folder, creating a new record dimension if one does not exist. The record dimension is often the time dimension, for example if you have a set of netCDF files, with each one representing some spatial field at a given timestep. If appropriate, you can rename this record dimension to something more useful using the ncrename utility (another utility in the NCO package).
Renaming dimensions
ncrename -d record,time merged.nc
The -d flag specifies that we are going to rename the dimension in the netcdf file., from “record” to “time”. There are also flags to rename other attributes, see the ncrename manual page
Removing degenerate dimensions
To remove degenerate dimensions (by averaging over the dimension to be removed):
ncwa -a dim_name input.nc output.nc
ncwa (“Weighted average”) will average variables over the specified dimension. If our dimension is degenerate (dim = 1), then this is effectively a way to remove that dimension without changing any of the variable data (Since it is averaging the variable over 1).
Adding a new variable
If we need to add a new variable, this can be done with ncap2 (ncap is deprecated).
ncap2 -s'new_dim[$new_dim]=1234'
Note that this will add a single value of the time variable: 1234.
Changing a variable to vary over a newly added dimension
If we add a new dimension, the existing variables will not automatically be functions of this new dimension. So if we were to add a time dimension, we need to recreate our variables to remap over this new dimension (assuming this is correct and appropriate for that particular variable/dimension combination.)
ncap2 -s 'Var_new[$dim1, $dim2, $new_dim3]=Var_old' input.nc output.nc
Further nco utilities
The available utilities with nco are:
The NCO utilities are
- ncap2 - arithmetic processor
- ncatted - attribute editor
- ncbo - binary operator
- ncdiff - differencer
- ncea - ensemble averager
- ncecat - ensemble concatenator
- ncflint - file interpolator
- ncks - kitchen sink (extract, cut, paste, print data)
- ncpdq - permute dimensions quickly
- ncra - running averager
- ncrcat - record concatenator
- ncrename - renamer
- ncwa - weighted averager
CDO (Climate Data Operators)
This is an equally capable set of netCDF tools written by the Max Planck Institute for Meteorology.

